CUPED
Experiment setup
goal is to estimate the delta = mean(Y_c) - mean(Y_t)
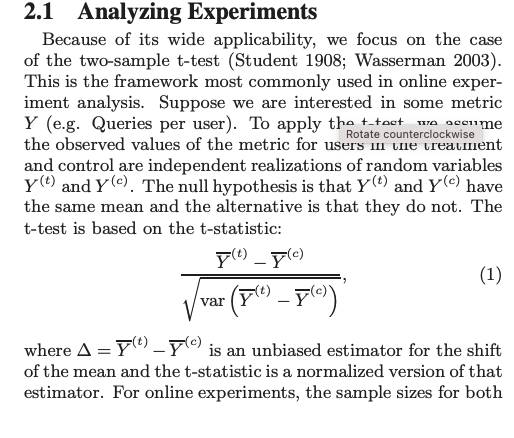
We want this estimate to have small varaince and to be unbiased.
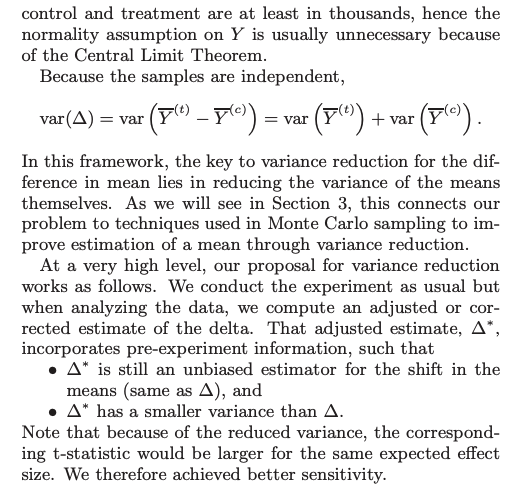
In this framework, the key to variance reduction for the difference in mean lies in reducing the variance of the means themselves.
What is CUPED?
CUPED is a well-established variance reduction technique for experiments paper
Unlike methods like outlier removal or winsorization, it doesn’t sacrifice any data integrity.
It can be used together with winsorization, which enhances its effectiveness.
It requires pre-experimentation data - which we generally have in Bunsen.
For a rather unpredictable event like ad clicks, we get ~5% reduction in standard deviation (which is ~10% reduction in variance, and experiment run time). For something like sessions, that 5% can increase to something like 30%.
Predicted uncertainty is not uncertainty.
So the variance in your experiment metric should only take into account the unpredictable part of your measurement. If you predict that:
- guv A will have 5 sessions during the experiment, and
- guv B will have 20 sessions,
And when you actually measure the sessions, you get:
- guv A has 4 sessions (-1 from prediction)
- guv B has 21 sessions (+1 from prediction),
Then the variance, or the uncertainty, in your measurement can be calculated on the residuals of your predictions, rather than on the measurements themselves. This gives var([-1, +1]), rather than var([4, 21]), which is a great reduction.
Control Variates
From paper:

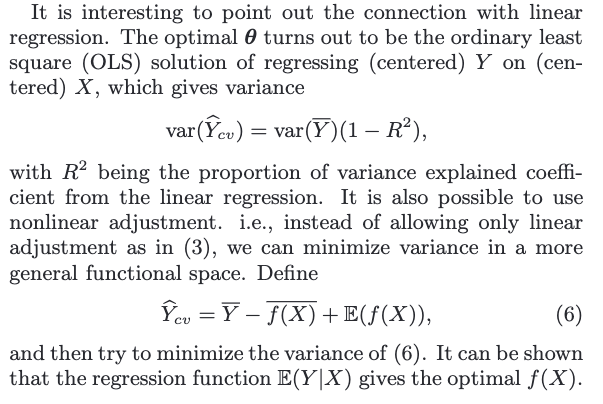
Control Variates in online experiments
The difficulty of applying it boils down to finding a control variate that is highly correlated with and at the same time has known .

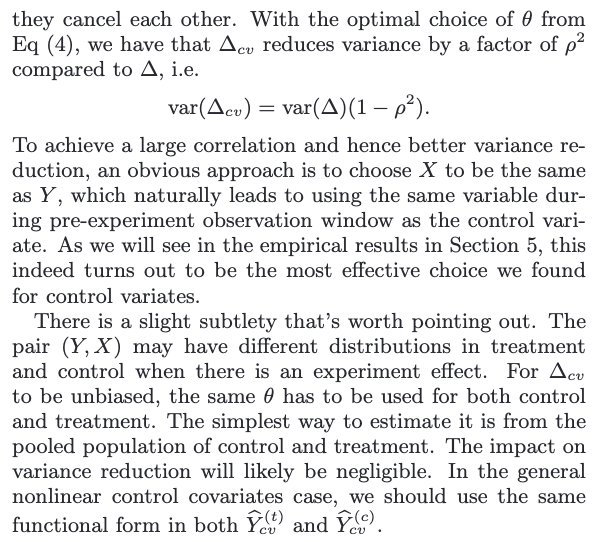
cv stands for control varaites
Control Variates vs Stratification
See Stratification
These are two techniques that both utilize covariates to achieve variance reduction. The stratification approach uses the covariates to construct strata while the control variates approach uses them as regression variables.
The former uses discrete (or discretized) covariates, whereas control variates seem more naturally to be continuous variables.
Control Variates is an extension of stratification, where the covariate can be an indicator variable showing the belonging of a sample to a stratum.
While these two techniques are well connected mathematically, they provide different insights into understanding why and how to achieve variance reduction. The stratification formulation has a nice analogy with mixture models, where each stratum is one component of the mixture model. Stratification is equivalent to separating samples according to their component memberships, effectively removing the between-component variance and achieving a reduced variance. A better covariate is hence the one that can better classify the samples and align with their underlying structure. On the other hand, the control variates formulation quantifies the amount of variance reduction as a function of the correlation between the covariates and the variable itself. It is mathematically simpler and more elegant. Clearly, a better covariate should be the one with larger (absolute) correlation.
CUPED IN PRACTICE
A simple yet effective way to implement CUPED is to use the same variable from the pre-experiment period as the covariate. You need to have pre-experiment data (have not applied treatment), not good when you work with new users.
The more correlated covariate with the target the larger variance reduction.
Across a large class of metrics, our results consistently showed that using the same variable from the pre-experiment period as the covariate tends to give the best variance reduction. In addition, the lengths of the pre-experiment and the experiment periods also play a role. Given the same pre-experiment period, extending the length of the experiment does not necessarily improve the variance reduction rate. On the other hand, a longer pre-period tends to give a higher reduction for the same experiment period.
- margarida's implementation
-- winsorization 90th %tile + CUPED
WITH bucketed_assignment_log AS (
-- macro assign_cohorts start
SELECT cohort_id,
user_id_encid,
MIN(first_assignment_time) - INTERVAL '5 SECOND' AS first_assignment_time
FROM dl_bunsen_filtered.assignment_log_metric_analysis_v2
WHERE experiment_run_id = 11052
AND (is_bot = FALSE OR is_bot IS NULL)
AND first_assignment_time BETWEEN '2023-07-25' -- wildcards are timestamp strings, e.g. '2022-07-28 12:00:00'
AND '2023-09-25' -- note that the single quotes are required
GROUP BY 1, 2
-- macro assign_cohorts end
),
connections_data AS (SELECT connections.user_id_encid,
connections.object_id,
datapipe_timestamp AS event_time
FROM dl_bunsen_mad.connections_sessionized_bot_labeled AS connections
WHERE TIMESTAMP 'epoch' + datapipe_event_time / 1000000 * INTERVAL '1 second' BETWEEN '2023-07-25'
AND '2023-09-25'
AND connections.dt BETWEEN (DATE_TRUNC('day', '2023-07-25'::TIMESTAMP) - INTERVAL '1 DAY')
AND (DATE_TRUNC('day', '2023-09-25'::TIMESTAMP) + INTERVAL '1 DAY')
AND connection_type IN ('photo_uploaded')
AND is_bot = 'false'
GROUP BY 1, 2, 3),
sum_per_day_per_user AS (SELECT cohort_id,
bucketed_assignment_log.user_id_encid,
first_assignment_time,
COUNT(DISTINCT object_id) AS sum_per_day_per_user
FROM bucketed_assignment_log
INNER JOIN connections_data
ON bucketed_assignment_log.user_id_encid = connections_data.user_id_encid
AND
connections_data.event_time BETWEEN bucketed_assignment_log.first_assignment_time
AND '2023-09-25'
GROUP BY 1, 2, 3),
percentile_reviews AS (SELECT user_id_encid,
sum_per_day_per_user,
PERCENTILE_CONT(0.90) WITHIN GROUP (ORDER BY sum_per_day_per_user)
OVER () AS percentile_90
FROM sum_per_day_per_user),
censored_photos AS (SELECT user_id_encid,
CASE
WHEN sum_per_day_per_user <= percentile_90 THEN sum_per_day_per_user
ELSE percentile_90
END AS photos_censored
FROM percentile_reviews),
sum_per_user AS (SELECT cohort_id,
user_id_encid,
first_assignment_time,
COALESCE(photos_censored, 0) /
(DATEDIFF(SEC,
TO_DATE('2023-07-25', 'YYYY-MM-DD'),
TO_DATE('2023-09-25', 'YYYY-MM-DD')) / 86400.
)::FLOAT AS avg_sum_per_user
FROM bucketed_assignment_log
LEFT JOIN censored_photos
USING (user_id_encid)),
get_theta AS (SELECT cohort_id,
user_id_encid,
X,
Y,
avg_X,
avg_Y,
var_X,
SUM((X - avg_X) * (Y - avg_Y) / (N - 1)) OVER () AS cov_XY,
cov_XY / var_X AS theta
FROM (SELECT cohort_id,
sum_per_user.user_id_encid,
COALESCE(num_photos_in_last_year, 0) AS X,
avg_sum_per_user AS Y,
COUNT(*) OVER () AS N,
AVG(X) OVER () AS avg_X,
AVG(Y) OVER () AS avg_Y,
VARIANCE(X) OVER () AS var_X
FROM sum_per_user
LEFT JOIN data_science.contribution_summary
ON sum_per_user.user_id_encid = contribution_summary.user_id_encid
AND contribution_summary.dt =
(SELECT MAX(dt) FROM data_science.contribution_summary)) AS aux
GROUP BY cohort_id,
user_id_encid,
X,
Y,
avg_X,
var_X,
avg_Y,
N),
get_Y_cuped AS (SELECT cohort_id,
user_id_encid,
Y - theta * (X - avg_X) AS Y_cuped
FROM get_theta
GROUP BY cohort_id,
user_id_encid,
X,
Y,
avg_X,
theta)
SELECT cohort_id,
COUNT(user_id_encid) AS sample_size,
AVG(Y_cuped) AS metric_value,
STDDEV(Y_cuped) AS standard_deviation
FROM get_Y_cuped
GROUP BY cohort_id
ORDER BY cohort_id;